diff options
Diffstat (limited to 'README.md')
| -rw-r--r-- | README.md | 19 |
1 files changed, 10 insertions, 9 deletions
diff --git a/README.md b/README.md index 2b3ab5d..c54de0e 100644 --- a/README.md +++ b/README.md @@ -61,33 +61,34 @@ pyhegp --help # How to use ## Simple data sharing - +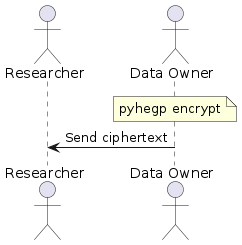 -In this simple scenario, there is only one data owner and they wish to share their encrypted data with a researcher. The data owner encrypts their data with: +In this simple scenario, there is only one data owner and they wish to share their encrypted data with a researcher. The data owner encrypts their genotype and phenotype data with: ``` -pyhegp encrypt genotype.tsv +pyhegp encrypt genotype.tsv phenotype.tsv ``` -They then send the encrypted data `genotype.tsv.hegp` to the researcher. Note that data sharing is carried out-of-band and is outside the scope of `pyhegp`. +They then send the encrypted `genotype.tsv.hegp` and `phenotype.tsv.hegp` to the researcher. Note that data sharing is carried out-of-band and is outside the scope of `pyhegp`. ## Joint/federated analysis with many data owners - +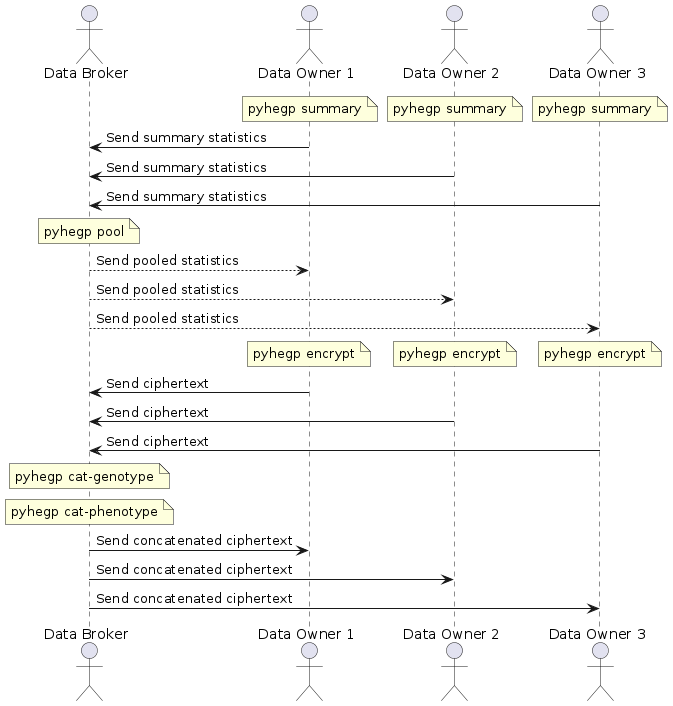 Data owners generate summary statistics for their data. ``` pyhegp summary genotype.tsv -o summary ``` -They share this with the data broker who pools it to compute the summary statistics of the complete dataset. +They share this with the data broker who pools it to compute the summary statistics of the complete dataset. Any SNPs not common to all summaries will be dropped. ``` pyhegp pool -o complete-summary summary1 summary2 ... ``` -The data broker shares these summary statistics with the data owners. The data owners standardize their data using these summary statistics, and encrypt their data using a random key. +The data broker shares these summary statistics with the data owners. The data owners standardize their data using these summary statistics, and encrypt their genotype and phenotype data using a random key. Any SNPs not in `complete-summary` or have a zero standard deviation are dropped. SNPs with a zero standard deviation have no discriminatory power in the analysis. ``` -pyhegp encrypt -s complete-summary genotype.tsv +pyhegp encrypt -s complete-summary genotype.tsv phenotype.tsv ``` -Finally, the data owners share the encrypted data `genotype.tsv.hegp` with the broker who concatenates it and shares it with all parties. +Finally, the data owners share the encrypted `genotype.tsv.hegp` and `phenotype.tsv.hegp` with the broker who concatenates it and shares it with all parties. ``` pyhegp cat-genotype -o complete-genotype.tsv.hegp genotype1.tsv.hegp genotype2.tsv.hegp ... +pyhegp cat-phenotype -o complete-phenotype.tsv.hegp phenotype1.tsv.hegp phenotype2.tsv.hegp ... ``` Note that all data sharing is carried out-of-band and is outside the scope of `pyhegp`. |
