diff options
| -rw-r--r-- | .guix/hsmice-test.scm | 205 | ||||
| -rw-r--r-- | .guix/pyhegp-package.scm | 15 | ||||
| -rw-r--r-- | .guix/readme-images.scm | 36 | ||||
| -rw-r--r-- | README.md | 8 | ||||
| -rwxr-xr-x | doc/generate-images.sh | 4 | ||||
| -rw-r--r-- | doc/joint-workflow.png | bin | 27569 -> 0 bytes | |||
| -rw-r--r-- | doc/joint-workflow.uml | 23 | ||||
| -rw-r--r-- | doc/simple-workflow.png | bin | 7260 -> 0 bytes | |||
| -rw-r--r-- | doc/simple-workflow.uml | 5 |
9 files changed, 203 insertions, 93 deletions
diff --git a/.guix/hsmice-test.scm b/.guix/hsmice-test.scm index 9137dc6..1982f65 100644 --- a/.guix/hsmice-test.scm +++ b/.guix/hsmice-test.scm @@ -66,83 +66,136 @@ genome-wide association study} library for R.") (license license:gpl3+))) -(define test-profile - (profile - (content (packages->manifest (list gzip tar pyhegp - python python-click python-pandas - r r-dplyr r-genio - r-mixed-model-gwas r-purrr - r-qqman r-readr r-stringr - r-tibble r-tidyr))))) - -(define wrangle-script - (local-file "../e2e-tests/hsmice/wrangle.r")) - -(define gwas-script - (local-file "../e2e-tests/hsmice/gwas.r")) - -(define check-qtl-script - (local-file "../e2e-tests/hsmice/check-qtl.py")) - -(define hsmice-test-gexp - (with-imported-modules '((guix build utils)) - #~(begin - (use-modules (guix build utils)) - - (mkdir #$output) - (set-path-environment-variable - "PATH" '("/bin") '(#$test-profile)) - (set-path-environment-variable - "GUIX_PYTHONPATH" - '(#$(string-append "/lib/python" - (version-major+minor (package-version python)) - "/site-packages")) - '(#$test-profile)) - (set-path-environment-variable - "R_LIBS_SITE" '("/site-library") '(#$test-profile)) - (invoke "tar" "-xvf" #$hsmice-data - "./HSmice/1_QTL_data/") - (invoke "Rscript" #$wrangle-script "HSmice/1_QTL_data" ".") - - ;; GWAS on plaintext - (invoke "Rscript" #$gwas-script - "genotype.tsv" "phenotype.tsv" - (string-append #$output "/plaintext-pvalues")) - (copy-file "Rplots.pdf" (string-append #$output "/plaintext-manhattan.pdf")) - - ;; GWAS with simple ciphertext data sharing - (invoke "pyhegp" "encrypt" "genotype.tsv" "phenotype.tsv") - (invoke "Rscript" #$gwas-script - "genotype.tsv.hegp" "phenotype.tsv.hegp" - (string-append #$output "/ciphertext-pvalues")) - (copy-file "Rplots.pdf" - (string-append #$output "/ciphertext-manhattan.pdf")) - - ;; Joint federated GWAS - (invoke "pyhegp" "summary" "genotype1.tsv" "-o" "summary1") - (invoke "pyhegp" "summary" "genotype2.tsv" "-o" "summary2") - (invoke "pyhegp" "pool" "-o" "complete-summary" "summary1" "summary2") - (invoke "pyhegp" "encrypt" "-s" "complete-summary" "genotype1.tsv" "phenotype1.tsv") - (invoke "pyhegp" "encrypt" "-s" "complete-summary" "genotype2.tsv" "phenotype2.tsv") - (invoke "pyhegp" "cat-genotype" "-o" "complete-genotype.tsv.hegp" - "genotype1.tsv.hegp" "genotype2.tsv.hegp") - (invoke "pyhegp" "cat-phenotype" "-o" "complete-phenotype.tsv.hegp" - "phenotype1.tsv.hegp" "phenotype2.tsv.hegp") - (invoke "Rscript" #$gwas-script - "complete-genotype.tsv.hegp" "complete-phenotype.tsv.hegp" - (string-append #$output "/federated-ciphertext-pvalues")) - (copy-file "Rplots.pdf" - (string-append #$output "/federated-ciphertext-manhattan.pdf")) - - ;; Check that the QTL is where it should be. - (for-each (lambda (pvalues-file) - (invoke "python3" #$check-qtl-script - (string-append #$output "/" pvalues-file))) - (list "plaintext-pvalues" - "ciphertext-pvalues" - "federated-ciphertext-pvalues"))))) +(define hsmice-wrangled-gexp + (let ((script-profile (profile + (content (packages->manifest + (list gzip tar r r-dplyr r-genio + r-purrr r-readr r-tibble r-tidyr)))))) + (with-imported-modules '((guix build utils)) + #~(begin + (use-modules (guix build utils)) + + (mkdir #$output) + (set-path-environment-variable + "PATH" '("bin") '(#$script-profile)) + (set-path-environment-variable + "R_LIBS_SITE" '("site-library") '(#$script-profile)) + (invoke "tar" "-xvf" #$hsmice-data + "./HSmice/1_QTL_data/") + (invoke "Rscript" + #$(local-file "../e2e-tests/hsmice/wrangle.r") + "HSmice/1_QTL_data" #$output))))) + +(define hsmice-wrangled + (computed-file "hsmice-wrangled" hsmice-wrangled-gexp)) + +(define hsmice-ciphertext-gexp + (let ((script-profile (profile + (content (packages->manifest (list pyhegp)))))) + (with-imported-modules '((guix build utils)) + #~(begin + (use-modules (guix build utils) + (srfi srfi-26)) + + (mkdir #$output) + (set-path-environment-variable + "PATH" '("bin") '(#$script-profile)) + (for-each (cut install-file <> (getcwd)) + (find-files #$hsmice-wrangled "\\.tsv$")) + ;; Simple data sharing workflow + (invoke "pyhegp" "encrypt" "genotype.tsv" "phenotype.tsv") + ;; Joint/federated analysis workflow + (invoke "pyhegp" "summary" "genotype1.tsv" "-o" "summary1") + (invoke "pyhegp" "summary" "genotype2.tsv" "-o" "summary2") + (invoke "pyhegp" "pool" "-o" "complete-summary" "summary1" "summary2") + (invoke "pyhegp" "encrypt" "-s" "complete-summary" "genotype1.tsv" "phenotype1.tsv") + (invoke "pyhegp" "encrypt" "-s" "complete-summary" "genotype2.tsv" "phenotype2.tsv") + (invoke "pyhegp" "cat-genotype" "-o" "complete-genotype.tsv.hegp" + "genotype1.tsv.hegp" "genotype2.tsv.hegp") + (invoke "pyhegp" "cat-phenotype" "-o" "complete-phenotype.tsv.hegp" + "phenotype1.tsv.hegp" "phenotype2.tsv.hegp") + (for-each (cut install-file <> #$output) + (find-files (getcwd) "\\.tsv.hegp$")))))) + +(define hsmice-ciphertext + (computed-file "hsmice-ciphertext" hsmice-ciphertext-gexp)) + +(define hsmice-r-mixed-model-gwas-gexp + (let ((gwas-script (local-file "../e2e-tests/hsmice/gwas.r")) + (script-profile (profile + (content (packages->manifest + (list r r-dplyr r-mixed-model-gwas + r-qqman r-readr r-stringr + r-tibble r-tidyr)))))) + (with-imported-modules '((guix build utils)) + #~(begin + (use-modules (guix build utils)) + + (mkdir #$output) + (set-path-environment-variable + "PATH" '("bin") '(#$script-profile)) + (set-path-environment-variable + "R_LIBS_SITE" '("site-library") '(#$script-profile)) + + ;; GWAS on plaintext + (invoke "Rscript" #$gwas-script + #$(file-append hsmice-wrangled "/genotype.tsv") + #$(file-append hsmice-wrangled "/phenotype.tsv") + (string-append #$output "/plaintext-pvalues")) + (copy-file "Rplots.pdf" (string-append #$output "/plaintext-manhattan.pdf")) + + ;; GWAS with simple ciphertext data sharing + (invoke "Rscript" #$gwas-script + #$(file-append hsmice-ciphertext "/genotype.tsv.hegp") + #$(file-append hsmice-ciphertext "/phenotype.tsv.hegp") + (string-append #$output "/ciphertext-pvalues")) + (copy-file "Rplots.pdf" + (string-append #$output "/ciphertext-manhattan.pdf")) + + ;; Joint federated GWAS + (invoke "Rscript" #$gwas-script + #$(file-append hsmice-ciphertext "/complete-genotype.tsv.hegp") + #$(file-append hsmice-ciphertext "/complete-phenotype.tsv.hegp") + (string-append #$output "/federated-ciphertext-pvalues")) + (copy-file "Rplots.pdf" + (string-append #$output "/federated-ciphertext-manhattan.pdf")))))) + +(define hsmice-r-mixed-model-gwas + (computed-file "hsmice-r-mixed-model-gwas" hsmice-r-mixed-model-gwas-gexp)) + +(define hsmice-qtl-checked-gexp + (let ((script-profile (profile + (content (packages->manifest + (list python python-pandas)))))) + (with-imported-modules '((guix build utils)) + #~(begin + (use-modules (guix build utils) + (srfi srfi-26)) + + (mkdir #$output) + (set-path-environment-variable + "PATH" '("bin") '(#$script-profile)) + (set-path-environment-variable + "GUIX_PYTHONPATH" + '(#$(string-append "lib/python" + (version-major+minor (package-version python)) + "/site-packages")) + '(#$script-profile)) + + ;; Check that the QTL is where it should be. + (for-each (cut invoke + "python3" + #$(local-file "../e2e-tests/hsmice/check-qtl.py") + <>) + (find-files #$hsmice-r-mixed-model-gwas + "\\-pvalues$")))))) + +(define hsmice-qtl-checked + (computed-file "hsmice-qtl-checked" hsmice-qtl-checked-gexp)) (define-public hsmice-test - (computed-file "hsmice-test" hsmice-test-gexp)) + (directory-union "hsmice-test" + (list hsmice-r-mixed-model-gwas + hsmice-qtl-checked))) hsmice-test diff --git a/.guix/pyhegp-package.scm b/.guix/pyhegp-package.scm index 20231cd..3eb3685 100644 --- a/.guix/pyhegp-package.scm +++ b/.guix/pyhegp-package.scm @@ -43,8 +43,19 @@ (source (local-file ".." "pyhegp-checkout" #:recursive? #t - #:select? (or (git-predicate (dirname (current-source-directory))) - (const #t)))) + #:select? (lambda (file stat) + ;; If .guix is included, changes + ;; to other files under .guix—such + ;; as the hsmice + ;; test—unnecessarily trigger a + ;; rebuild of pyhegp. This could + ;; be a nuisance when hacking on + ;; the test scripts. + (and (not (string-contains file "/.guix/")) + (not (string-contains file "/e2e-tests/")) + ((or (git-predicate (dirname (current-source-directory))) + (const #t)) + file stat))))) (build-system pyproject-build-system) (arguments (list #:phases diff --git a/.guix/readme-images.scm b/.guix/readme-images.scm new file mode 100644 index 0000000..5579fd7 --- /dev/null +++ b/.guix/readme-images.scm @@ -0,0 +1,36 @@ +;;; pyhegp --- Homomorphic encryption of genotypes and phenotypes +;;; Copyright © 2025 Arun Isaac <arunisaac@systemreboot.net> +;;; +;;; This file is part of pyhegp. +;;; +;;; pyhegp is free software: you can redistribute it and/or modify it +;;; under the terms of the GNU General Public License as published by +;;; the Free Software Foundation, either version 3 of the License, or +;;; (at your option) any later version. +;;; +;;; pyhegp is distributed in the hope that it will be useful, but +;;; WITHOUT ANY WARRANTY; without even the implied warranty of +;;; MERCHANTABILITY or FITNESS FOR A PARTICULAR PURPOSE. See the GNU +;;; General Public License for more details. +;;; +;;; You should have received a copy of the GNU General Public License +;;; along with pyhegp. If not, see <https://www.gnu.org/licenses/>. + +(define-module (readme-images) + #:use-module ((gnu packages uml) #:select (plantuml)) + #:use-module (guix gexp)) + +(define readme-images-gexp + (with-imported-modules '((guix build utils)) + #~(begin + (use-modules (guix build utils)) + + (invoke #$(file-append plantuml "/bin/plantuml") + #$(local-file "../doc/simple-workflow.uml") + #$(local-file "../doc/joint-workflow.uml") + "-o" #$output)))) + +(define-public readme-images + (computed-file "pyhegp-readme-images" readme-images-gexp)) + +readme-images diff --git a/README.md b/README.md index 399b39a..c54de0e 100644 --- a/README.md +++ b/README.md @@ -61,7 +61,7 @@ pyhegp --help # How to use ## Simple data sharing - +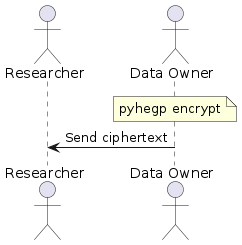 In this simple scenario, there is only one data owner and they wish to share their encrypted data with a researcher. The data owner encrypts their genotype and phenotype data with: ``` @@ -71,17 +71,17 @@ They then send the encrypted `genotype.tsv.hegp` and `phenotype.tsv.hegp` to the ## Joint/federated analysis with many data owners - +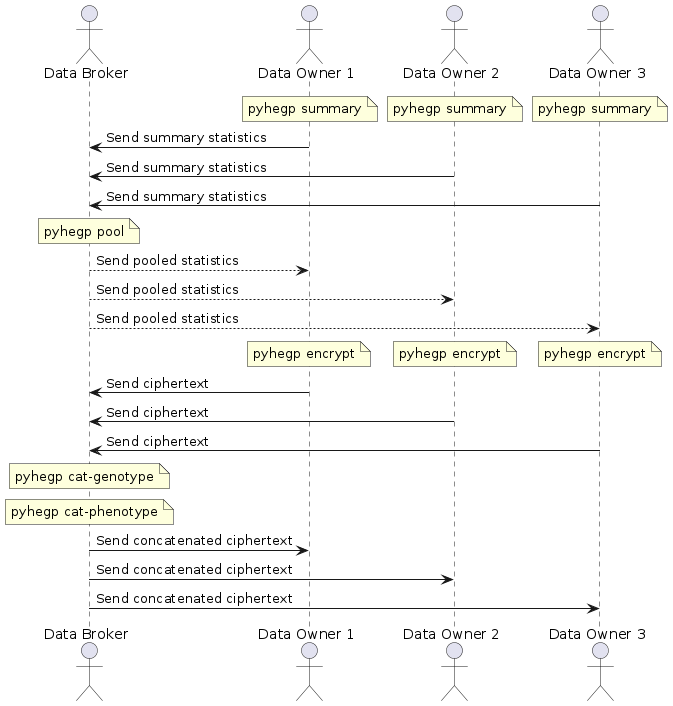 Data owners generate summary statistics for their data. ``` pyhegp summary genotype.tsv -o summary ``` -They share this with the data broker who pools it to compute the summary statistics of the complete dataset. +They share this with the data broker who pools it to compute the summary statistics of the complete dataset. Any SNPs not common to all summaries will be dropped. ``` pyhegp pool -o complete-summary summary1 summary2 ... ``` -The data broker shares these summary statistics with the data owners. The data owners standardize their data using these summary statistics, and encrypt their genotype and phenotype data using a random key. +The data broker shares these summary statistics with the data owners. The data owners standardize their data using these summary statistics, and encrypt their genotype and phenotype data using a random key. Any SNPs not in `complete-summary` or have a zero standard deviation are dropped. SNPs with a zero standard deviation have no discriminatory power in the analysis. ``` pyhegp encrypt -s complete-summary genotype.tsv phenotype.tsv ``` diff --git a/doc/generate-images.sh b/doc/generate-images.sh deleted file mode 100755 index 0950519..0000000 --- a/doc/generate-images.sh +++ /dev/null @@ -1,4 +0,0 @@ -#! /bin/sh - -cat simple-workflow.uml | guix shell plantuml -- plantuml -p > simple-workflow.png -cat joint-workflow.uml | guix shell plantuml -- plantuml -p > joint-workflow.png diff --git a/doc/joint-workflow.png b/doc/joint-workflow.png deleted file mode 100644 index b2ff1b2..0000000 --- a/doc/joint-workflow.png +++ /dev/null Binary files differdiff --git a/doc/joint-workflow.uml b/doc/joint-workflow.uml index 2d1542c..8488b79 100644 --- a/doc/joint-workflow.uml +++ b/doc/joint-workflow.uml @@ -1,16 +1,27 @@ +@startuml joint-workflow actor "Data Broker" as broker actor "Data Owner 1" as owner1 actor "Data Owner 2" as owner2 actor "Data Owner 3" as owner3 +note over owner1: pyhegp summary +/ note over owner2: pyhegp summary +/ note over owner3: pyhegp summary owner1 -> broker: Send summary statistics owner2 -> broker: Send summary statistics owner3 -> broker: Send summary statistics +note over broker: pyhegp pool broker --> owner1: Send pooled statistics broker --> owner2: Send pooled statistics broker --> owner3: Send pooled statistics -owner1 -> broker: Encrypt and share ciphertext -owner2 -> broker: Encrypt and share ciphertext -owner3 -> broker: Encrypt and share ciphertext -broker -> owner1: Share concatenated ciphertext -broker -> owner2: Share concatenated ciphertext -broker -> owner3: Share concatenated ciphertext +note over owner1: pyhegp encrypt +/ note over owner2: pyhegp encrypt +/ note over owner3: pyhegp encrypt +owner1 -> broker: Send ciphertext +owner2 -> broker: Send ciphertext +owner3 -> broker: Send ciphertext +note over broker: pyhegp cat-genotype +note over broker: pyhegp cat-phenotype +broker -> owner1: Send concatenated ciphertext +broker -> owner2: Send concatenated ciphertext +broker -> owner3: Send concatenated ciphertext +@enduml \ No newline at end of file diff --git a/doc/simple-workflow.png b/doc/simple-workflow.png deleted file mode 100644 index 5d9f01d..0000000 --- a/doc/simple-workflow.png +++ /dev/null Binary files differdiff --git a/doc/simple-workflow.uml b/doc/simple-workflow.uml index 02dc005..9a9d088 100644 --- a/doc/simple-workflow.uml +++ b/doc/simple-workflow.uml @@ -1,3 +1,6 @@ +@startuml simple-workflow actor "Researcher" as researcher actor "Data Owner" as owner -owner -> researcher: Encrypt and share ciphertext \ No newline at end of file +note over owner: pyhegp encrypt +owner -> researcher: Send ciphertext +@enduml |
