pyhegp is a Python library and CLI utility implementing homomorphic encryption of genotypes and phenotypes as described in - Private Genomes and Public SNPs: Homomorphic Encryption of Genotypes and Phenotypes for Shared Quantitative Genetics - Using encrypted genotypes and phenotypes for collaborative genomic analyses to maintain data confidentiality
Table of contents
- Install development version
- Using pip
- Using Guix
- How to use
- Simple data sharing
- Joint/federated analysis with many data owners
- Frequently asked questions (FAQ)
- File formats
- Run tests
- License
Install development version
The development version of pyhegp may be installed either using pip or using Guix.
Using pip
Create a virtual environment (optional)
In a new directory, create a python virtual environment and activate it.
mkdir pyhegp
cd pyhegp
python3 -m venv .venv
source .venv/bin/activate
Install pyhegp
Install the development version of pyhegp. If you are in a virtual environment, pyhegp will be installed in it. If you skipped the previous step, pyhegp will be installed in your user install directory (that's typically in your home directory).
pip install git+https://github.com/encryption4genetics/pyhegp
Using Guix
Put the following into a channels.scm file.
(use-modules (guix ci))
(list (channel
(name 'pyhegp)
(url "https://github.com/encryption4genetics/pyhegp")
(branch "main"))
(channel-with-substitutes-available %default-guix-channel
"https://ci.guix.gnu.org"))
Build a Guix profile using this channels.scm and activate it.
guix pull -C channels.scm -p pyhegp-profile
source ./pyhegp-profile/etc/profile
Drop into a shell where pyhegp is installed.
guix shell pyhegp
Now, you can use pyhegp.
pyhegp --help
How to use
Simple data sharing
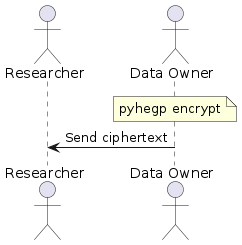
In this simple scenario, there is only one data owner and they wish to share their encrypted data with a researcher. The data owner encrypts their genotype and phenotype data with:
pyhegp encrypt genotype.tsv phenotype.tsv
They then send the encrypted genotype.tsv.hegp and phenotype.tsv.hegp to the researcher. Note that data sharing is carried out-of-band and is outside the scope of pyhegp.
Joint/federated analysis with many data owners
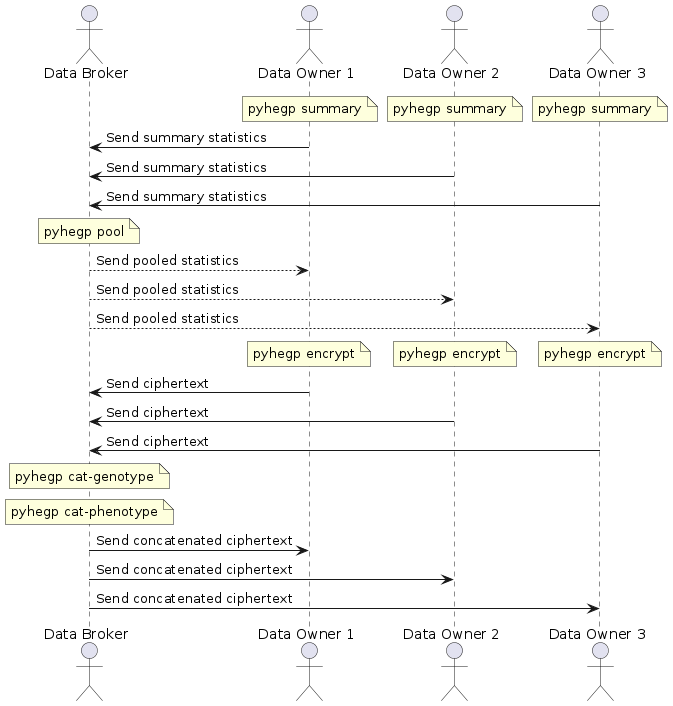
Data owners generate summary statistics for their data.
pyhegp summary genotype.tsv -o summary
They share this with the data broker who pools it to compute the summary statistics of the complete dataset. Any SNPs not common to all summaries will be dropped.
pyhegp pool -o complete-summary summary1 summary2 ...
The data broker shares these summary statistics with the data owners. The data owners standardize their data using these summary statistics, and encrypt their genotype and phenotype data using a random key. Any SNPs not in complete-summary or have a zero standard deviation are dropped. SNPs with a zero standard deviation have no discriminatory power in the analysis.
pyhegp encrypt -s complete-summary genotype.tsv phenotype.tsv
Finally, the data owners share the encrypted genotype.tsv.hegp and phenotype.tsv.hegp with the broker who concatenates it and shares it with all parties.
pyhegp cat-genotype -o complete-genotype.tsv.hegp genotype1.tsv.hegp genotype2.tsv.hegp ...
pyhegp cat-phenotype -o complete-phenotype.tsv.hegp phenotype1.tsv.hegp phenotype2.tsv.hegp ...
Note that all data sharing is carried out-of-band and is outside the scope of pyhegp.
Frequently asked questions (FAQ)
Does pyhegp standardize the data?
For each SNP, pyhegp standardizes the genotype (so the mean is zero and the standard deviation is one) during encryption. pyhegp does NOT standardize the phenotype.
File formats
See File formats for documentation of file formats used by pyhegp.
Run tests
Run the test suite using
python3 -m pytest
The test suite is not meant to be run by end users. It is meant to be run by developers when hacking on the code.
License
pyhegp is free software released under the terms of the GNU General Public License, either version 3 of the License, or (at your option) any later version.
